PyNAST 1.2.1 release, and moving the PyNAST blog
Hi all,
I’m happy to announce the PyNAST 1.2.1 release, available for download here. This is primarily a bug fix release, allowing PyNAST to make smarter decisions about where to store temporary files. This had been an issue for some PyNAST users working in cluster environments. If this hasn’t been an issue for you, it is not important to upgrade from PyNAST 1.2.0.
Also, this will be the last post on the PyNAST blog. All future news and announcements related to PyNAST will be posted to the QIIME blog, so if you’re not already subscribed there, you should sign up. The QIIME blog is a very low traffic list (typically less that one message per month). Merging these blogs will help me reduce my administrative burden so I can spend more time doing science and writing software.
Thanks for using PyNAST, and hope to see you on the QIIME blog!
Greg
PyNAST 1.1 released: better alignments and a 15x speed increase!
The PyNAST 1.1 release went live today with several notable improvements. The first thing users need to know is that PyNAST now requires PyCogent 1.4.1 (get it here) and uclust 1.1.579 (get it here). Also, BLAST is no longer required, but can optionally be used for the pairwise alignment step. Unless you have specific reasons for using it, for example if you expect that one or both of the ends of your reads may be problematic, I recommend sticking with the default pairwise aligner, which is now uclust. uclust used a speed-optimized Needleman-Wunsch alignment.
The list of features that were added differs somewhat from the slated features announced last week. Of the three major changes I mentioned, the only one that actually made it into this release was feature 1, the switch from BLAST for sequence searching and aligning, to uclust for searching and aligning. This gave some nice improvements to the alignment quality over using BLAST, so for the time-being I decided to hold off on features 2 and 3.
A major enhancement in PyNAST 1.1 is that an issue which lead to poor alignments, and thereby artificially long branches in phylogenetic trees, has been dealt with. It appears that differences in BLAST parameter settings were causing rare differences between NAST and PyNAST 1.0 alignments. Large gaps were being handled in different way between the two implementations: NAST would truncate the sequence to avoid a poor alignment, while PyNAST would try to align the region. By default, PyNAST 1.1 uses a global rather than local alignment, and as a consequence achieves better alignments in these cases. Interested users can check out this alignment — I recommend viewing this in a text editor (e.g., TextMate), turning off soft wrapping of lines, and jumping to around column 1637 to see where the difference in implementations begin to differ. As you can see, PyNAST 1.1 achieves the best alignment of the three with respect to the template sequence. This is also evident when comparing FastTree phylogenies built from lanemasked variants of the full alignments. The long branch in Figure 1a corresponds to the sequence presented in the alignment.
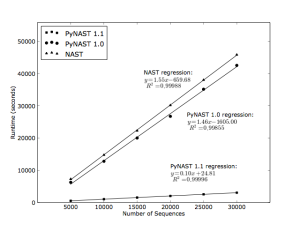
In addition to better alignments, a major improvement in PyNAST 1.1 is a decrease in runtime of approximately 15 times compared to PyNAST 1.0, when using the default parameters in both cases. I attribute this primarily to the switch from BLAST to uclust — thanks a lot to Robert Edgar, author of uclust, for all the help with making the switch to uclust. Our uclust wrappers now live in PyCogent (beginning with version 1.4.1), and are used extensively in PyNAST and QIIME. See Figure 2 for an illustration of the improvement in runtime when comparing PyNAST 1.1 to PyNAST 1.0 and the NAST command line application.
Finally, an important note for PyNAST’s microbial ecologist and QIIME users. While the resulting alignments — and therefore the derived phylogenetic trees — are improved in PyNAST 1.1, the difference has little or no affect on beta diversity conclusions. Figure3 illustrates this point by comparing PCoA plots which summarize unweighted Unifrac distances between samples in the QIIME tutorial data set, when alignments were constructed with PyNAST 1.0 (Figure 3a) and PyNAST 1.1 (Figure 3b). There is little difference between the two plots, and certainly no difference in the conclusions drawn from the figures.

Figure 3a: PCoA plots summarizing unweighted unifrac distances between samples in the QIIME tutorial data set. Alignments were generated with PyNAST 1.0.

Figure 3b: PCoA plots summarizing unweighted unifrac distances between samples in the QIIME tutorial data set. Alignments were generated with PyNAST 1.1.
Thanks to all users and collaborators for the helpful suggestions that went into PyNAST 1.1!
-Greg
Changes to PyNAST in preparation for PyNAST 1.1
The PyNAST code is undergoing significant changes this week in preparation for PyNAST 1.1, which I’m hoping to release by 2 April 2010. This message is primarily important for users of the PyNAST svn code.
In this release we’re no longer aiming to match the results of the original NAST web server and command line application, but rather to focus on getting better alignments. Users who want to match those results will always be able to use PyNAST 1.0.
PyNAST’s command line interface will mostly stay the same in PyNAST 1.1. The major changes are improvements to the underlying PyNAST algorithm. While I’m making these changes over the next few days, the svn code is likely to be somewhat unstable. I recommend that svn users go with one of the following strategies:
- stick with the svn version that you’re currently using until PyNAST 1.1 is released;
- use PyNAST 1.0 until PyNAST 1.1 is released;
- or, at least, be aware that PyNAST alignments may change from version to version over the next few days.
As always, the svn code is less stable than the release code. I do generally encourage users to work with the svn version if they can tolerate some instability. Your input helps the development team improve PyNAST by identifying issues prior to packing up releases.
Here’s a sneak peak of some of the new features:
Feature 1
Switch from default of searching and aligning with BLAST to searching and aligning with uclust. Because uclust uses k-mer searching and performs global alignments, this addresses findings from Schloss 2009 (briefly, that k-mer searching is better, and that global alignment works at least as well as local alignment for this application). Additionally, this will effectively remove the dependence on BLAST, which some users have trouble with, and should speed PyNAST up (possibly by orders of magnitude).
Feature 2
Support will be added for using a different pairwise alignment based on %id of the candidate and the best template match. By default, the uclust alignment will always be used, but this will leave the door open for (e.g.) aligning with structure-based aligners (like infernal) for more distant matches. Depending on how long other changes take, I may not add support for this to the command line interface in PyNAST 1.1, but I hope to at least add it to the API so it’s accessible via QIIME.
Feature 3
More informed choice of gaps positions to delete when decreasing the alignment length by incorporating conservation data. We’ve recently noticed some long-branch issues resulting from trees generated from PyNAST alignments. I think these occur when a bad choice is made for the gap to remove to reduce the length of the candidate sequence alignment.
My initial thought on how to achieve this is that we can allow users to (optionally) provide a lanemask file (like the greengenes lanemask). If provided, rather than simply removing the nearest gap, PyNAST can count the number of conserved positions (i.e., lanemask=1) that would be disrupted by different choices of gap positions to remove. It would then remove the gap that disturbs the fewest conserved positions (and default to the closest gap in the event of a tie). I plan to do some experimentation with this, and see if it would address some of the long branch results that people have shared with me recently.
Thanks!
Greg
PyNAST 1.0 released!
The first point release of PyNAST was released on 25 January 2009, and is available for download from sourceforge.
This release is likely of interest to users who are more interested in a stable release than in having access to the latest changes. Users who are interested in the latest changes can still check PyNAST out of our sourceforge SVN repository.